BIO-HELIX - DP001-0100
nanoTaq Hot-Start DNA Polymerase
Size: 100rxns (500units /100ul)
Bio-Helix nanoTaq DNA Polymerase was designed as one of enhanced hot start enzyme DNA Polymerases which provide the convenience and reliability toward your research destination. nanoTaq was engineered with nano technology complex, which is an innovative creation, different from the traditional methods of hot start enzymes made. The proven features of nanoTaq covers reactions at room temperature using the same protocol and cycling conditions as conventional Taq DNA polymerases, reducing nonspecific primer annealing, improving product yield and using for PCR products up to 5kb.
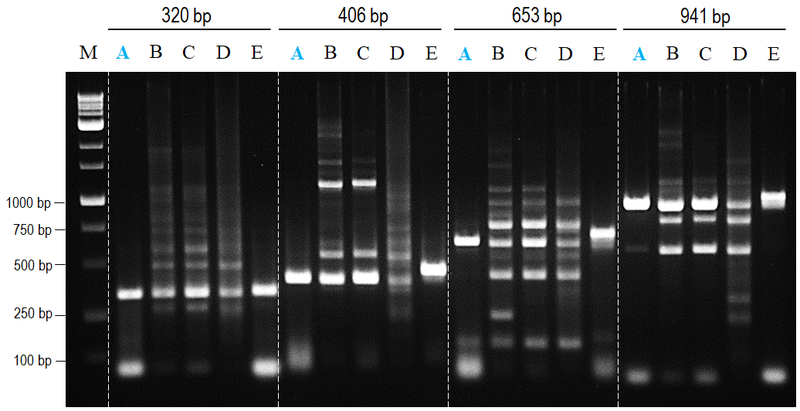
nanoTaq DNA polymerase provides both specificity and yield.
|
A |
nanoTaq |
|
B |
Thermo DreamTaq |
|
C |
Thermo DreamTaq HS |
|
D |
Roche KAPA2G HS |
|
E |
Invitrogen Platinum HS |
Figure 1. Comparison of non-specific amplifying effects to major suppliers.
All amplifications were performed in accordance with manufacturer’s instructions to amplify 320 to 941 bp amplicons from human genomic DNA. nanoTaq provided both higher specificity and yield products with least band shifting compared to other commercial antibody-mediated hot start polymerase.
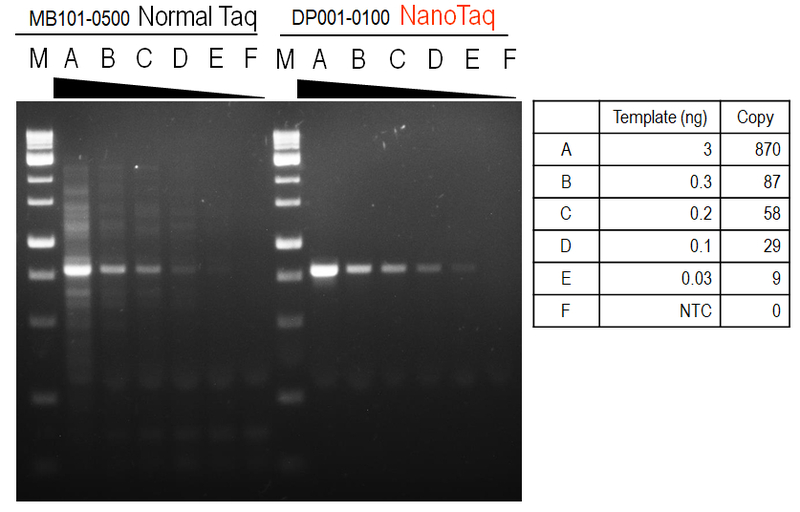
Figure 2. Sensitivity and reliable amplification from low amounts of input DNA.
Amplification of a 803 bp fragment from 3; 0.3; 0.2; 0.1; 0.03; 0(no template control) ng of human genomic DNA were amplified in 20μL PCR reactions using nanoTaq DNA Polymerase.
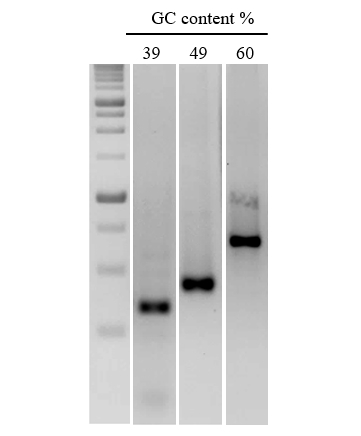
Figure 3. Efficient amplification of DNA sequences with a range of GC content.
A series of DNA fragments of increasing GC content were amplified from human gDNA. nanoTaq Hot-Start DNA Polymerase was used for targets with >50% GC.
*Thermo DreamTaq, Thermo DreamTaq HS and Invitrogen Platinum HS are registered trademarks of Thermo ScientificTM PierceTM. Roche KAPA2G HS is a registered trademark of Merck. The trademark holders are not affiliated with Bio-Helix Co., Ltd. and do not endorse these products.
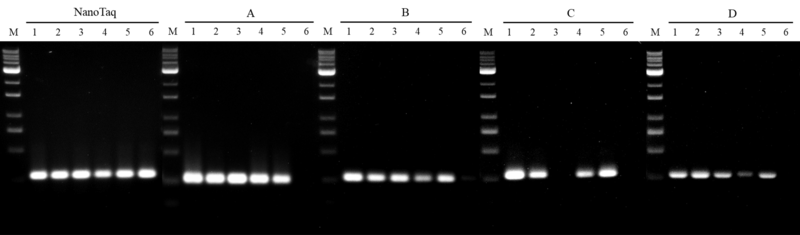
A. Thermo DreamTaq, Cat. EP0701
B. Thermo DreamTaq HS, Cat. EP1701
C. Roche KAPA2G HS, Cat. KK5523
D. Invitrogen Platinum HS, Cat. 10966-018
Figure 4. Inhibitor resistance on nanoTaq DNA polymerase
A 280 bp human genomic DNA target was conducted to compare with four leading competing DNA polymerases. All amplifications were performed in accordance with manufacturer’s instructions where the mixtures contain:
1. no inhibitor control,
2. bile salts with final concentration of 0.5 mg/mL,
3. heparin with final concentration of 0.5 unit/mL,
4. sodium citrate with final concentration of 150 μg/mL,
5. hemin with final concentration of 0.1 μg/mL and
6. humic acid with final concentration of 1 μg/mL.
The results show that nanoTaq has the best tolerance with the humic acid interference, when compared with major suppliers of the hot-start DNA polymerase.
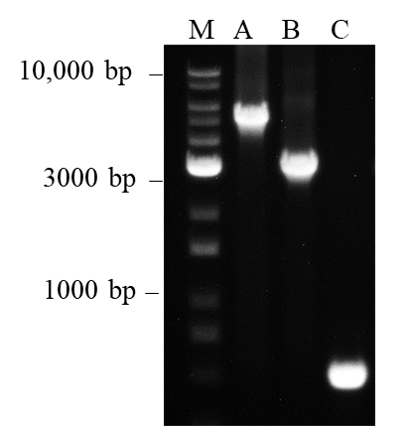
Figure 5. Amplification of long target DNA fragments
It is generally known that Taq DNA polymerase exhibits difficulty amplifying DNA fragment sizes larger than 1 kb. This is probably due to the polymerase’s error-prone amplification process which reduces the efficiency in amplifying longer target sizes. However, we have shown that nanoTaq is capable of amplifying Lane M: 1Kb plus DNA Ladder RTU (DM015-R500), Lane A: 5 kb, Lane B: 3 kb and Lane C: 0.5 kb Lambda DNA fragment sizes under the 3-min extension time at 72°C. Therefore, nanoTaq shows the optimal performance in applications with long PCR fragments.
► High tolerance aganist various reaction inhibitors
► Reduce nonspecific primer annealing
► Plays in efficient PCR amplification of GC-rich sequences
► Using the same protocol and cycling conditions as conventional Taq DNA polymerases
1. For each 20 μl reaction, assemble the following in a 0.2 ml PCR tube on ice just prior to use:
| Volume | Final Conc. | |
| DNA template | - μl | 3 ng |
| Forward primer, 5-10 μM | - μl | 0.1-0.5 μM |
| Reverse primer, 5-10 μM | - μl | 0.1-0.5 μM |
| dNTP Mix (10 mM each dATP, dCTP, dGTP, dTTP) | - μl | 200 µM |
| 10X PCR buffer | 2 μl | - |
| nanoTaq Hot-Start DNA Polymerase | -1 μl | - |
| PCR Grade Water | Add to 20 μl | - |
| Total volume | 20 μl | - |
2. Mix gently. If necessary, centrifuge briefly and cap tubes.
3. Place them into Thermocycler and process for 30-35 cycles as follows:
| Initial Denaturation | 3 min at 95°C | |
| Denaturation | 30 sec | 30-35 cycles |
| Annealing | 30 sec at the proper annealing temperature | |
| Extension | 1 min at 72°C | |
| Final extension | 5 min at 72°C |
Note: Optimal conditions for amplification will vary depending on the primers and thermal cycler used. It may be necessary to optimize the system for individual primers, template, and thermal cycler.
* Template:
Purified high quality DNA is needed for a success PCR reaction. The final concentration of cDNA template please refer to “Reaction Setup”.
* Storage Buffer:
The enzyme is supplied in a storage buffer consisting of 20 mM Tris-HCl (pH 8.0), 0.1 mM EDTA, 1 mM DTT, 50% (v/v) glycerol, and 1% Triton X-100.
* Unit Definition:
One unit is defined as the amount of enzyme that will catalyze the incorporation of 10 nmol of dNTP into acid- insoluble form in 30 min at 74°C in a reaction containing 25 mM TAPS (tris-[hydroxymethyl]-methyl-amino- propane-sulfonic acid, sodium salt), pH 9.3 at 25°C, 50 mM KCl, 2 mM MgCl2, 1 mM β-mercaptoethanol, 0.2 mM dATP, dGTP, and dTTP, 0.1 μM [α-32P] dCTP, and activated salmon sperm DNA.
| Name | Download |
|---|---|
| Safety Data Sheet|SDS | DP001_NanoTaq_Hot-Start_DNA_polymerase_SDS_20210713.pdf |
| PROTOCOL | Bio-Helix_DP001_Protocol_V7_20241114.pdf |